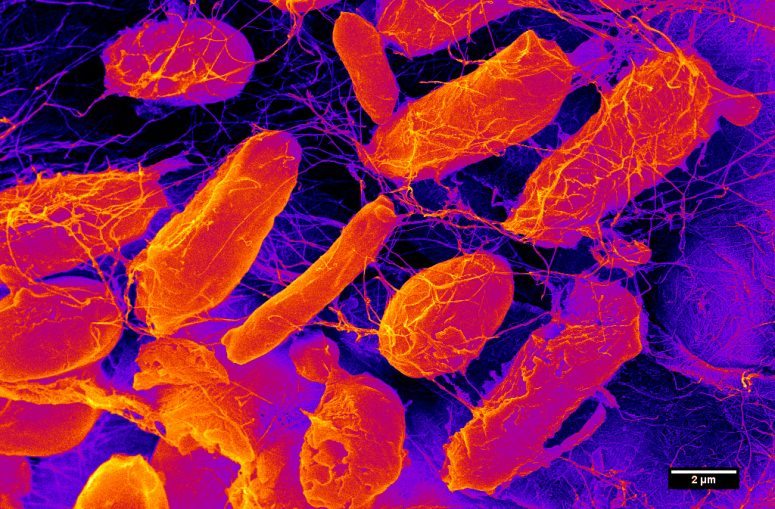
Since the 17th century, when Antonie van Leeuwenhoek first observed microorganisms through the lens of a rudimentary microscope, humans have slowly come to appreciate that ours is a germy world.
Through the ages with van Leeuwenhoek, Louis Pasteur and Robert Koch, the 19th century scientist who found that microorganisms could cause disease, human awareness of the microbial world and its importance has expanded to help underpin critical medical and agricultural discoveries, such as antibiotics and nitrogen-fixing bacteria, as well as to make us masters of the organisms that enrich our lives and diets through ordinary bread and wine.
In more recent years, microbes have proved their worth through things like polymerase chain reaction, a now common method to amplify DNA in the lab, in forensics and in medicine. The process depends on an enzyme from a bacterium retrieved from a hot spring in Yellowstone National Park in the 1960s and fuels billions of dollars in economic activity annually. In addition, the re-tasking of a natural microbial immune system, CRISPR, has enabled precise genome editing from microbes to plants and animals.
Now, seeking to further harness microbes’ many uses, the federal government has launched the National Microbiome Initiative (NMI) to “foster the integrated study of microbiomes across different ecosystems.”
Microbiomes are defined as communities of microorganisms that live on or in people (and other animals), plants, soil, oceans and the atmosphere.
The initiative will put us in a position to better understand microbes in context and how they work, explains University of Wisconsin—Madison bacteriology Professor Timothy Donohue. Its resources will depend much on the next federal budget, but various funding agencies as well as private organizations have committed to finding new support to enhance budgets for work related to the initiative.
“It is a scientific fact that ‘microbes touch everything,’ from the food we eat, the air we breathe and the water we drink,” says Donohue. “They are the master chemists of the universe, responsible for the world around us, and are intimately linked to the future health and evolution of the planet and its inhabitants.”
UW-Madison, Donohue argues, has both strengths and challenges in terms of capitalizing on the initiative. “Wisconsin has a history of and currently benefits by having many of the global thought leaders in microbiology,” he says. “However, to realize the potential of the NMI, we can benefit from leaders from disciplines who traditionally have not worked in the microbial sciences.”

In addition to strong programs in microbiology, he lists Wisconsin’s disciplinary breadth and long history of interdisciplinary research as assets. Researchers in fields such as chemistry, engineering, business and ethics will work to demystify microbiomes, how they function and how they might be exploited for the benefit of human health, food and energy production, environmental remediation and basic discovery.
A challenge, he says, will be aligning the technologies necessary to be successful in what is sure to be a competitive market for both capital and intellectual resources.
“There are major technology areas of need in this arena, including data analysis, modeling, imaging, technology development, biodesign, materials science, biomanufacturing and others,” Donohue says. “Some of these are common to this initiative and others, so there is intense national and international competition for leaders in these emerging areas.”
Trina McMahon, a UW–Madison professor of bacteriology and civil and environmental engineering, is cautiously optimistic about the national microbiome push, if for no other reason than it puts a spotlight on a corner of biology that is only now wending its way into public consciousness.
“The buzz is awesome,” she says. “Bringing the spotlight to the microbiome generally lets people know it’s not just about the gut microbiome,” an area that has received notoriety due mostly to its implications for human health.
McMahon studies microbial ecology in freshwater systems such as Lake Mendota and in the sludge processed at wastewater treatment plants. Her group is particularly interested in organisms that store phosphorus, a chemical nutrient and pollutant that helps spur the lake’s epic algae blooms.
At UW–Madison, exploration of the microbiome is occurring in many different labs and contexts, ranging from surveys of the microbiomes of the bat wing, copepods and Lake Michigan algae to the gut microbiomes of the Wisconsin high school class of 1957 as part of the Wisconsin Longitudinal Study (WLS).
Bacteriologist Federico Rey, who helps lead the WLS microbiome effort, is excited about the national push, but is naturally concerned about the competition. “I think it is really exciting to see how much interest there is in this field. Funding for this kind of research is going up, but it is nerve-wracking to see how much money other universities are putting into it. How is the University of Wisconsin going to compete?”
Wisconsin’s strengths, he says, reside in breadth of expertise, “a collaborative spirit and fantastic students.” But to be successful in a big way will require bringing all of those things and associated technologies — rapid gene sequencing, bioinformatics, computational resources and biologists — together to solve big problems.

The microbiome can also be a powerful teaching opportunity. Melissa Christopherson, a faculty associate in the Department of Bacteriology, took her microbiology capstone students on a tour of the microbiome of the human mouth this past semester, comparing the oral microbiomes of student athletes to see if there were differences in the microbial communities in elite athletes compared to others and if diet had an influence on the microbial cast of characters. The project’s findings are now being prepared for publication.
“One of the aims of this initiative is education,” says Christopherson, who was on hand for the May 13 White House summit where the initiative was announced by Jo Handelsman, a former UW–Madison professor of bacteriology and now associate director for science in the White House Office of Science and Technology Policy.
“There are only one or two examples of a course like this around the country,” Christopherson notes. “The students in this course got their money’s worth. They worked their butts off. A project like this is a way to summarize a lot of the things they’ve learned.”
The initiative rests on the growing array of technologies that make the sequencing and analysis of genetic material cheap and easy, says civil and environmental engineering Professor Dan Noguera. Most microorganisms can’t be cultured in the lab, but they can be cracked open and their genetic material can be plumbed with growing speed and accuracy.
“We’re able to do things we weren’t able to do 10 or 12 years ago,” says Noguera, who uses a Madison sewage treatment plant as a laboratory. “There are some very sophisticated tools and models, but only a few people are good at using them and interpreting them, so the hope is there will be some synergy.”
One potential upshot of the initiative, he observes, is the opportunity for the development of centers on the scale of the Department of Energy’s Great Lakes Bioenergy Research Center, a collaboration of UW–Madison and Michigan State University whose mission is to develop the next generation of biofuels.