University of Illinois Researchers create first model to directly connect photosynthesis and crop growth
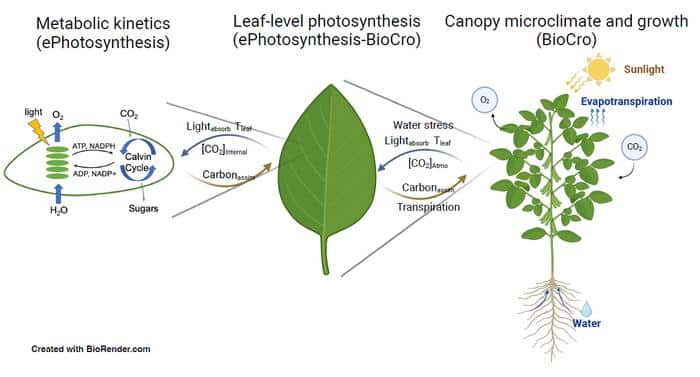
A team from the University of Illinois (UI) developed a modeling framework that connects enzyme activity related to photosynthesis to crop yield, marking the first time such a model directly links dynamic photosynthetic pathways to crop growth.
“A previous model coupled data from the genetic, metabolic, and leaf levels, but we needed to connect the metabolic level to the greater canopy level,” UI Matthews Group postdoctoral researcher Yufeng He said in a recent UI news release. “The new model allows us to examine how changes in enzyme activities can affect yield by better connecting the environmental variation experienced by the crops in the field to the metabolic processes.”
This research is part of the Realizing Increased Photosynthetic Efficiency (RIPE) project, an international initiative aimed at engineering crops to be more productive by improving photosynthesis. RIPE’s work received support from the Bill & Melinda Gates Foundation, Foundation for Food & Agriculture Research, U.K. Foreign, Commonwealth & Development Office, and Bill & Melinda Gates Agricultural Innovations (Gates Ag One).
In a study published in in silico Plants, He and colleagues demonstrated how their model enhances scientists’ ability to accurately simulate crop growth. Previously, simulations had to assume photosynthesis and associated enzyme activities were in a steady state, an unrealistic condition for crops in the field.
“Plants don’t exist in a stable environment. We can use this work to study the sensitivity of enzymes under different environmental conditions,” said Megan Matthews, principal investigator for the RIPE Project and assistant professor in civil and environmental engineering at Illinois. “The model will allow us to see which photosynthetic enzymes are limiting in different environments, and how they can lead to yield gain under long-term climatic conditions.”
The model’s success in identifying limiting enzymes and connecting them to expected yield is due to its detailed representation of dynamic enzyme reactions, moving beyond simplified steady-state representations. It also allows exploration of non-steady-state photosynthetic responses, such as those triggered by shade from cloud cover or wind movement. Previous dynamic models failed to scale up to crop growth at the field level.
“Scaling up from the metabolite level to the field level represents a pivotal advancement towards achieving a more accurate simulation of photosynthesis and crop growth,” said He. “Our coupled model holds promise for enhancing our comprehension of enzymatic dynamics and crop physiology. Furthermore, its future applications will assist in improving crop management strategies, fostering sustainable agriculture practices, and bolstering food security amidst global challenges.”
He and Matthews hope that their open-source model, available on GitHub, will help researchers better understand and predict how specific enzymes affect crop growth and yield in various environments.