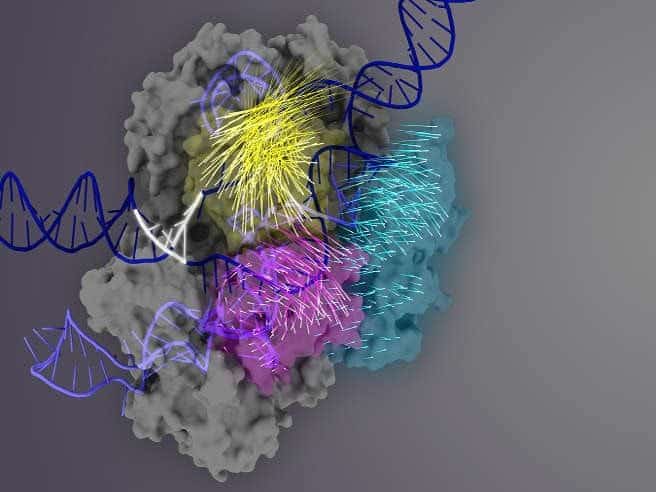
Scientists at the University of California, Berkeley and Massachusetts General Hospital have identified a key region within the Cas9 protein that governs how accurately CRISPR-Cas9 homes in on a target DNA sequence, and have tweaked it to produce a hyper-accurate gene editor with the lowest level of off-target cutting to date.
The protein domain the researchers identified as a master controller of DNA cutting is an obvious target for re-engineering to improve accuracy even further, the researchers say. This approach should help scientists customize variants of Cas9 – the protein that binds and cuts DNA – to minimize the chance that CRISPR-Cas9 will edit DNA at the wrong place, a key consideration when doing gene therapy in humans.
One strategy to achieve improved accuracy is to create mutations in the governing protein domain, called REC3, and see which ones improve accuracy without impacting the efficiency of on-target cutting.
“We have found that even minor alterations in the REC3 domain of Cas9 affect the differential between on- and off-target editing, which suggests that this domain is an obvious candidate for in-depth mutagenesis to improve targeting specificity. As an extension of this work, one could perform a more unbiased mutagenesis within REC3 than the targeted mutations we have made,” said co-first author Janice Chen, a graduate student in the lab of Jennifer Doudna, who co-invented the CRISPR-Cas9 gene-editing tool.
Co-first authors Chen, Yavuz Dagdas and Benjamin Kleinstiver, and their colleagues at UC Berkeley, Massachusetts General Hospital and Harvard University report their results online today in advance of publication in the journal Nature.
Hyper-accurate Cas9
Since 2012, when Doudna, a professor of molecular and cell biology and of chemistry and a Howard Hughes Medical Institute investigator at UC Berkeley, and colleague Emmanuelle Charpentier at the Max Planck Institute for Infection Biology repurposed the Cas9 protein to create a cheap, precise and easy-to-use gene editor, researchers have sought to lessen the chances of off-target editing. While improved fidelity benefits basic research, it is absolutely critical when editing genes for clinical applications, since any off-target DNA cutting could disable key genes and lead to permanent, unexpected side effects.
Within the last two years, two teams engineered highly accurate Cas9 proteins – an enhanced specificity one called eSpCas9(1.1) and a high-fidelity one called SpCas9-HF1 – and Chen and Doudna sought to learn why they cut with higher specificity than the wild-type Cas9 protein from Streptococcus pyogenes used widely today.
Currently, researchers using CRISPR-Cas9 create a single-guide RNA (sgRNA) – an RNA molecule that includes a chain of 20 ribonucleic acids that complements a specific 20-nucleic-acid DNA sequence they want to target — and attach it to Cas9. This guide RNA allows Cas9 to home in on the complementary DNA, bind to it and cut the double stranded helix. But the Cas9-sgRNA complex can also bind to DNA that doesn’t exactly match, leading to undesirable off-target cutting.
In 2015, Doudna’s lab discovered a conformational switch of Cas9 that is activated when the RNA guide and DNA target match. They found that only when the RNA and DNA match closely does the 3D structure of Cas9, in particular the conformation of the HNH nuclease domain, change and activate the scissors of Cas9. However, the process responsible for sensing the nucleic acids upstream of the conformational switch remained unknown.
In the current study, Chen and Dagdas used a technique called single-molecule FRET (Förster resonance energy transfer) to precisely measure how the various protein domains in the Cas9-sgRNA protein complex – in particular REC3, REC2 and HNH – move when the complex binds to DNA.
Maintaining efficiency while upping fidelity
They first determined that the specificity benefits conferred by eSpCas9(1.1) and SpCas9-HF1 could be explained by the fact that the threshold for the HNH conformational switch was much higher for these Cas9 variants than for the wild-type Cas9 protein, making the eSpCas9(1.1) and SpCas9-HF1 variants less likely to activate the scissors when bound to an off-target sequence.
Next, they uncovered that the REC3 domain is responsible for sensing the accuracy of target binding, which then signals the outward rotation of the REC2 domain to open a path for the HNH nuclease domain, activating the scissors. This active conformation of Cas9 is then able to cleave both strands of the target DNA.
Chen, Dagdas and Kleinstiver then showed that by mutating parts of REC3, it is possible to change the specificity of the Cas9 protein so that the HNH nuclease is not activated unless the guide RNA and target DNA match is very close. They were able to engineer an improved hyper-accurate Cas9, dubbed HypaCas9, that retains its on-target efficiency but is slightly better at discriminating between on- and off-target sites in human cells.
“If you mutate certain amino acid residues in REC3, you can tweak the balance between Cas9 on-target activity and improved specificity; we were able to find the sweet spot where there is sufficient activity at the intended target but also a large reduction in off-target events,” Chen said.
By continuing to explore the relationships between structure, function and dynamics of Cas9, Doudna and her team hope to further engineer the protein with exquisite sensitivity to reliably and efficiently perform a variety of genetic alterations.
Co-authors of the paper include Keith Joung, a professor at Harvard and Mass General, whose lab engineered the high-fidelity Cas9, SpCas9-HF1; Ahmet Yildiz, a UC Berkeley associate professor of molecular and cell biology and of physics; graduate student Lucas Harrington and former postdoc Samuel Sternberg of UC Berkeley; and Moira Welch and Alexander Sousa of Mass General.
The work was supported by the National Institutes of Health (GM094522, GM118773, R35 GM118158) and the National Science Foundation (MCB-1617028).