Abstract
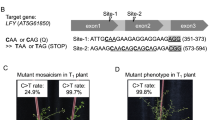
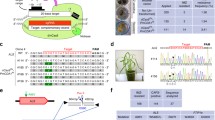
The CRISPR/Cas9-mediated base editing technology can efficiently generate point mutations in the genome without introducing a double-strand break (DSB) or supplying a DNA donor template for homology-directed repair (HDR). In this study, adenine base editors (ABEs) were used for rapid generation of precise point mutations in two distinct genes, OsWSL5, and OsZEBRA3 (Z3), in both rice protoplasts and regenerated plants. The precisely engineered point mutations were stably inherited to subsequent generations. These single nucleotide alterations resulted in single amino acid changes and associated wsl5 and z3 phenotypes as evidenced by white stripe leaf and light green/dark green leaf pattern, respectively. Through selfing and genetic segregation, transgene-free, base edited wsl5 and z3 mutants were obtained in a short period of time. We noticed a novel mutation (V540A) in Z3 locus could also mimic the phenotype of Z3 mutation (S542P). Furthermore, we observed unexpected non- A/G or T/C mutations in the ABE editing window in a few of the edited plants. The ABE vectors and the method from this study could be used to simultaneously generate point mutations in multiple target genes in a single transformation and serve as a useful base editing tool for crop improvement as well as basic studies in plant biology.



Similar content being viewed by others
References
Alseth I, Dalhus B, Bjørås M (2014) Inosine in DNA and RNA. Curr Opin Genet Dev 26:116–123
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Gaudelli NM, Packer MS, Liu DR, Komor AC, Bryson DI, Badran AH, Rees HA, Gaudelli NM (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551:464–471
Herraiz T, Galisteo J (2018) Nitrosative deamination of 2′-deoxyguanosine and DNA by nitrite, and antinitrosating activity of β-carboline alkaloids and antioxidants. Food Chem Toxicol 112:282–289
Hua K, Tao X, Yuan F, Wang D, Zhu JK (2018) Precise A•T to G•C base editing in the rice genome. Mol Plant 11:627–630
Hua K, Tao X, Zhu JK (2019a) Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J 17(2):499–504
Hua K, Tao X, Han P, Wang R, Zhu JK (2019b) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant 12:1003–1014
Huang TK, Puchta H (2019) CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep 38:443–453
Huang S, Weigel D, Beachy RN, Li J (2016) A proposed regulatory framework for genome-edited crops. Nat Genet 48:109–111
Ishizaki T (2016) CRISPR/Cas9 in rice can induce new mutations in later generations, leading to chimerism and unpredicted segregation of the targeted mutation. Mol Breed 36:165. https://doi.org/10.1007/s11032-016-0591-7
Kang B-C, Choi M, Ryu J, Woo JW, Yun J-Y, Kim J-S, Shin Y, Kim S-T (2018) Precision genome engineering through adenine base editing in plants. Nat Plants 4:427–431
Kim SH, Kwon CT, Song G, Koh HJ, An G, Paek NC (2018) The rice zebra3 (z3) mutation disrupts citrate distribution and produces transverse dark-green/green variegation in mature leaves. Rice 11:1–15
Kim HS, Jeong YK, Hur JK, Kim JS, Bae S (2019) Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol 37(10):1145–1148
Komor AC, Kim YB, Packer MS, Zuris JA, David R (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603):420–424
Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10:526–529
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:1–9. https://doi.org/10.1186/s13059-018-1443-z
Liu X, Lan J, Huang Y, Cao P, Zhou C, Ren Y, He N, Liu S, Tian Y, Nguyen T, Jiang L, Wan J (2018) WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot 69:3949–3961
Malina A, Cameron CJF, Robert F, Blanchette M, Dostie J, Pelletier J (2015) PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun 6:4–9. https://doi.org/10.1038/ncomms10124
Mei C, Qi M, Sheng G, Yang Y (2007) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant-Microb Interact 19:1127–1137
Molla KA, Yang Y (2019a) Predicting CRISPR/Cas-induced mutations for precise genome editing. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2019.08.002
Molla KA, Yang Y (2019b) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Molla KA, Debnath AB, Ganie SA, Mondal TK (2015) Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L). BMC Plant Biol. https://doi.org/10.1186/s12870-015-0498-1
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305):729
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H (2019) Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant 12:1015–1026
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40:1–12
Xie K, Yang Y (2013) RNA-Guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci 112:3570–3575
Xie X, Zeng D, Liu Y-G, Li G, Ma X, Zhu Q (2017) CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant 10:1246–1249
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:1–10. https://doi.org/10.1038/srep11491
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H (2018) Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11:631–634
Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y (2019) Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant 12:1027–1036
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35:438–440
Acknowledgements
Kutubuddin Molla would like to acknowledge the United States-India Educational Foundation (USIEF), New Delhi and the US Department of State for the Fulbright-Nehru Postdoctoral Fellowship. This work was supported by National Science Foundation Plant Genome Research Program Grant No. 1740874 and the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project #PEN04659 and Accession #1016432 to Yinong Yang.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Molla, K.A., Shih, J. & Yang, Y. Single-nucleotide editing for zebra3 and wsl5 phenotypes in rice using CRISPR/Cas9-mediated adenine base editors. aBIOTECH 1, 106–118 (2020). https://doi.org/10.1007/s42994-020-00018-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-020-00018-x